I’m Francisco Javier Guzmán Vega
PhD Candidate in Bioscience
Bioinformatician ~ Biologist ~ Developer


Trained as a Biotechnology Engineer, passionate in Bioinformatics and Computer Science. I am committed to the application of Bioinformatics and AI to solve problems in research and healthcare. I am an independent learner who’s obsessed with achieving a deep understanding of the problems around me and how to solve them.

About me
Born in Puebla, Mexico. Lived in different places all around the country. Studied my B.Sc. in Monterrey, and then took flight and landed in the Kingdom of Saudi Arabia, where I live currently.
Skills and interests

Python / 7 years
Usage of HPC cluster / 5 years
Linux / 7 years
Protein variant interpretation / 7 years
R / Basic knowledge
Biology / since B.Sc.
Web development / Basic knowledge
Statistics / Not my specialty but not too bad
Current Projects
Automated protein variant annotation
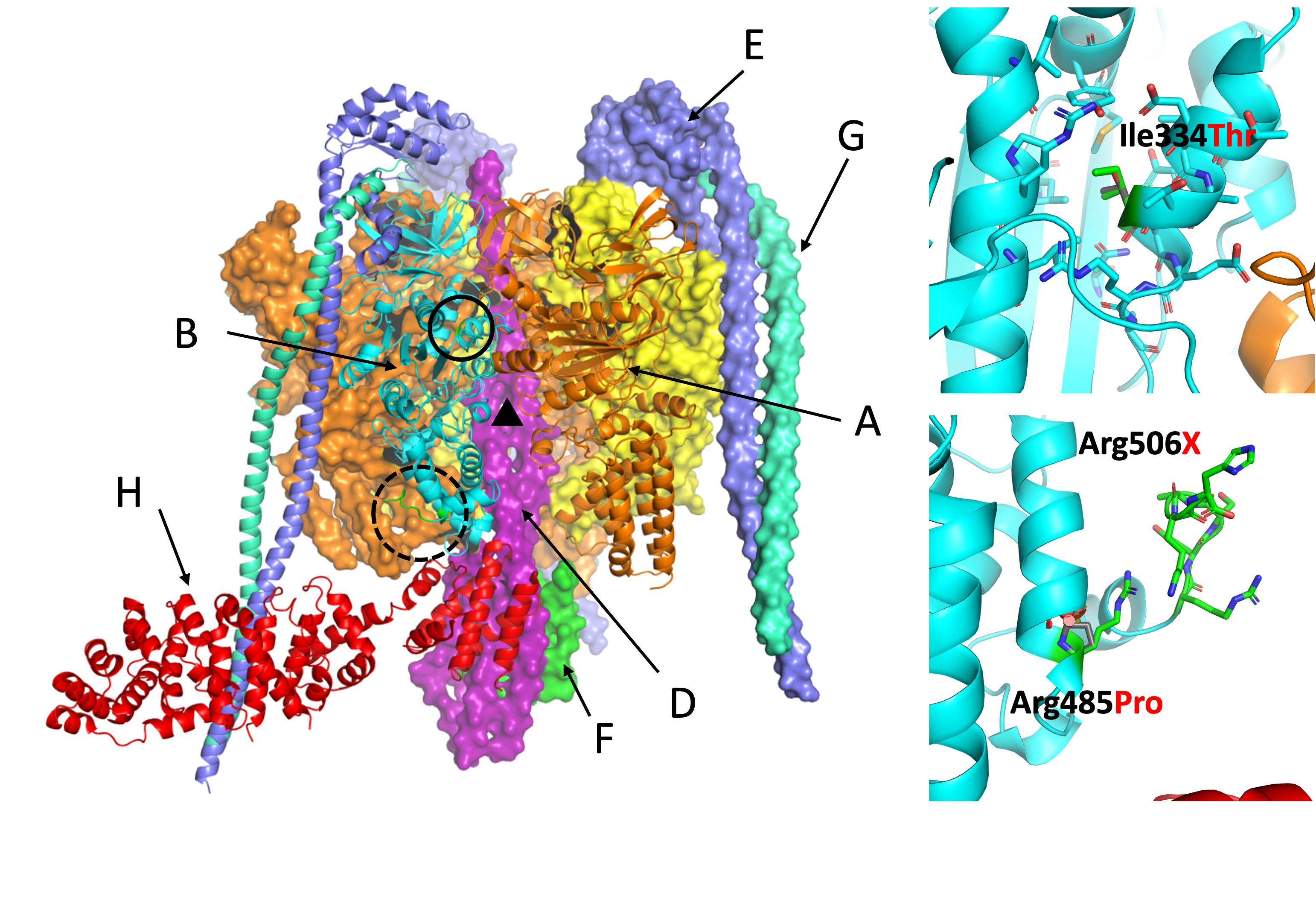
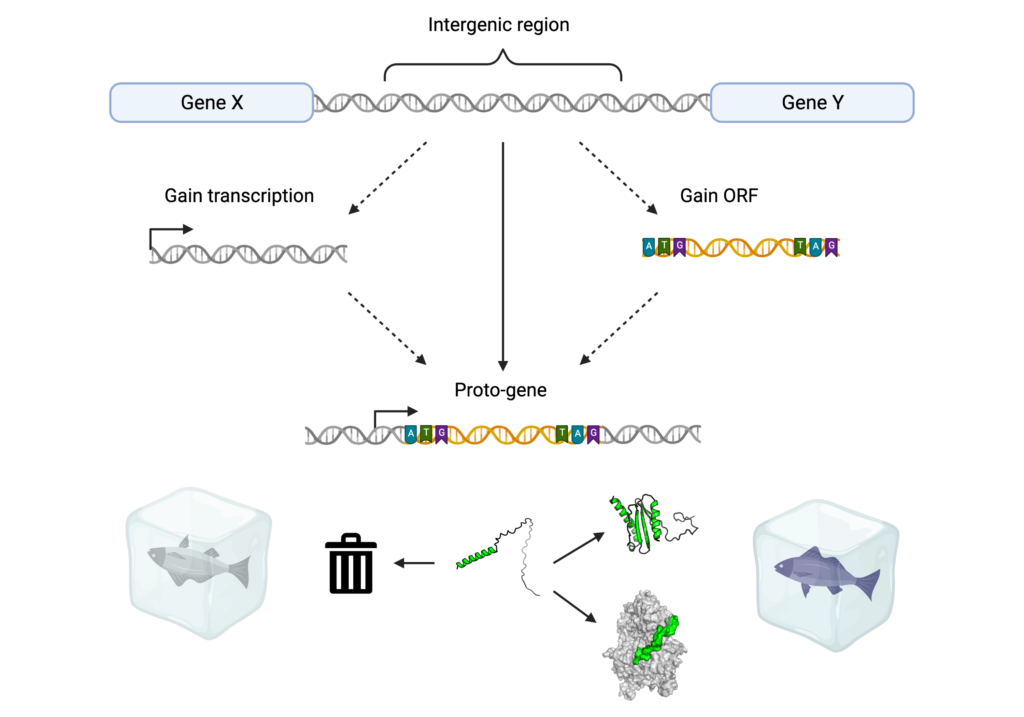
Fold and function of de novo proteins
Software and tools developed
AlphaFold-Ibex
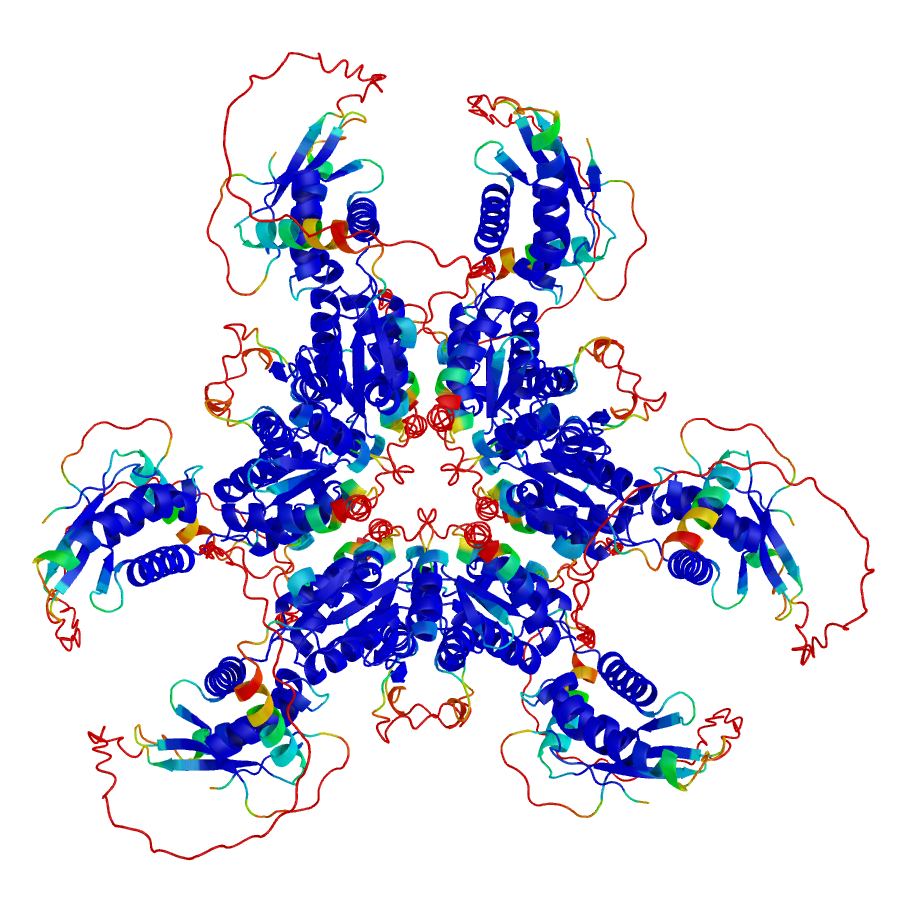
AlphaCRV
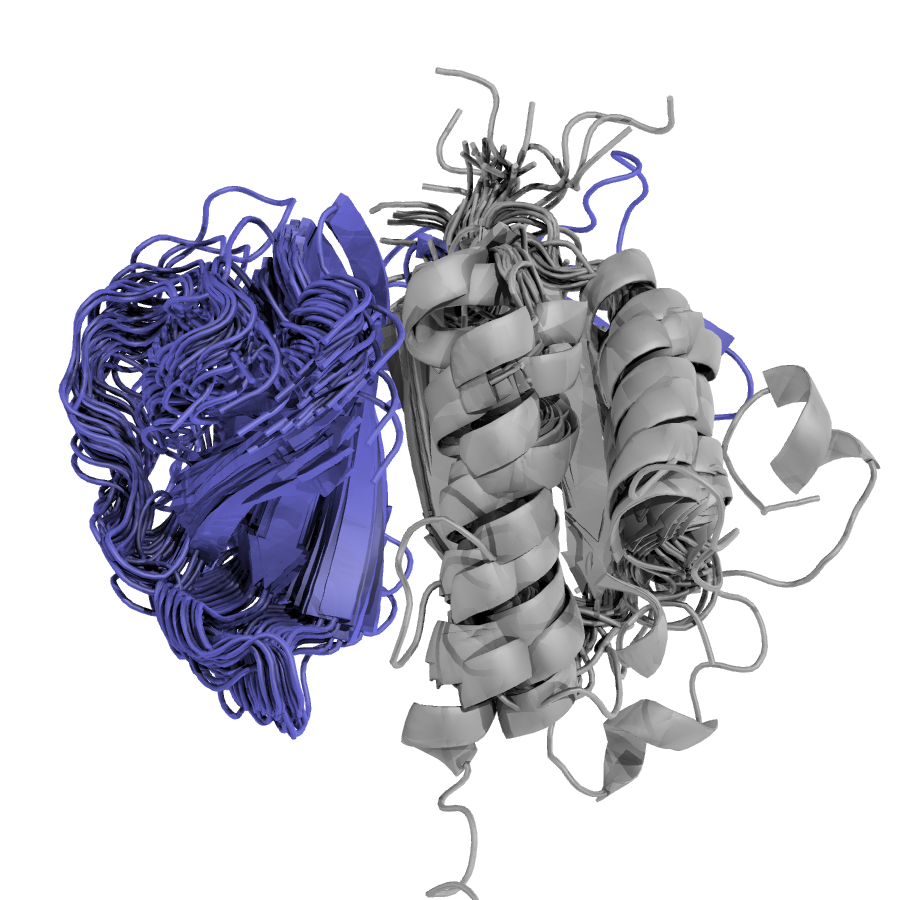
Multiprot
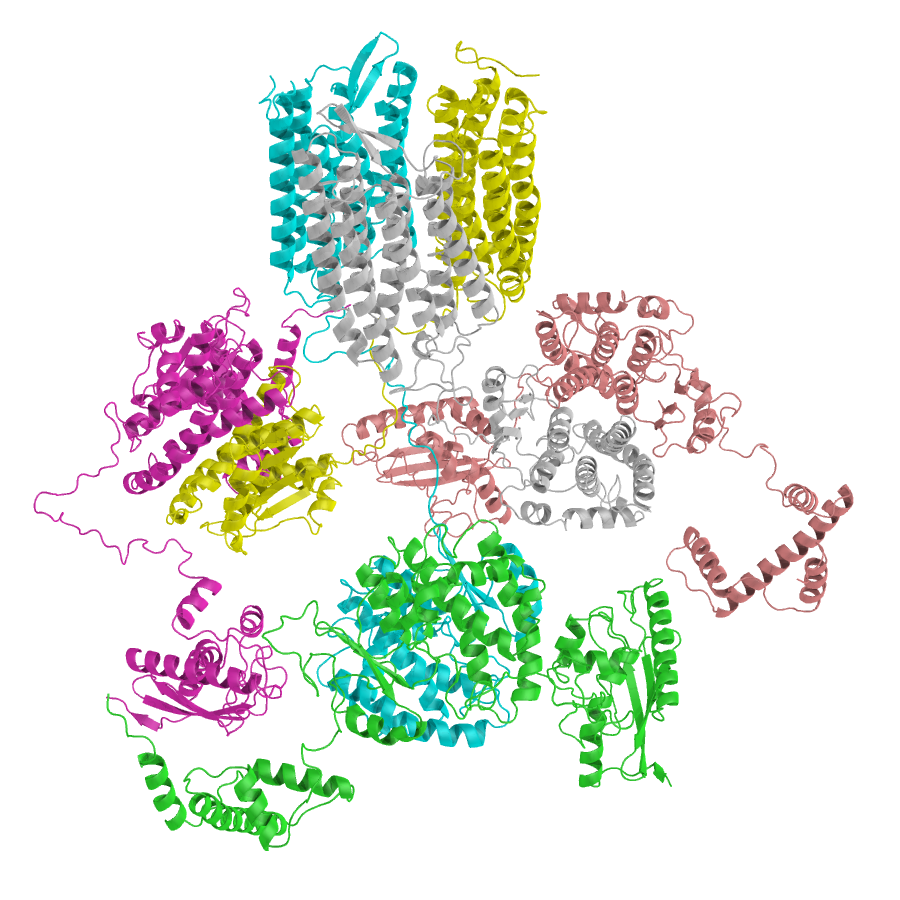
Recent Publications
AlphaCRV: A Pipeline for Identifying Accurate Binder Topologies in Mass-Modeling with AlphaFold
Guzmán-Vega, F. J. & Arold, S. T. (2024). bioRxiv. doi: https://doi.org/10.1101/2024.02.07.578780
I developed a pipeline to find the true binding topologies in proteome-wide protein-protein interaction screens.
Leveraging AI Advances and Online Tools for Structure-Based Variant Analysis
Guzmán-Vega, F. J., González-Álvarez, A. C., Peña-Guerra, K. A., Cardona-Londoño, K. J., & Arold, S. T. (2023). Current Protocols, 3, e857. https://doi.org/10.1002/cpz1.857.
I led the design and drafting of a protocol to help clinicians and researchers interpret protein variants from a structural perspective.
Novel Homozygous Variants of SLC13A5 Expand the Functional Heterogeneity of a Homogeneous Syndrome of Early Infantile Epileptic Encephalopathy.
Alsemari, A., Guzmán-Vega, F. J., Meyer, B. F., Arold, S. T. (2024) Pediatric Neurology, 151, 68-72. https://doi.org/10.1016/j.pediatrneurol.2023.10.005.
I modeled the 3D structure of the SLC13A5 dimer and evaluated disease-associated variants that strongly suggested an impairment of the sodium/citrate transport of SLC13A5.
An open-source, automated, and cost-effective platform for COVID-19 diagnosis and rapid portable genomic surveillance using nanopore sequencing.
Ramos-Mandujano, G., Grünberg, R., Zhang, Y., Bi, C., Guzmán-Vega, F. J. et al. (2023). Sci Rep, 13, 20349. https://doi.org/10.1038/s41598-023-47190-w.
I helped implement an automated RNA extraction protocol on a Tecan EVO-200 liquid handling robot.
Rapid Evolution of Plastic-degrading Enzymes Prevalent in the Global Ocean.
Alam, I., Aalismail, N., Martin, C., Kamau, A., Guzmán-Vega, F. J. et al. (2020). bioRxiv. doi: https://doi.org/10.1101/2020.09.07.285692.
I modeled the 3D structure of putative oceanic PETases, and designed a scoring system to rank their potential efficiency compared to the PETase from Ideonella sakaiensis.

Get in touch!
francisco.guzmanvega@kaust.edu.sa
www.linkedin.com/in/guzmanfj